Leukemogenesis
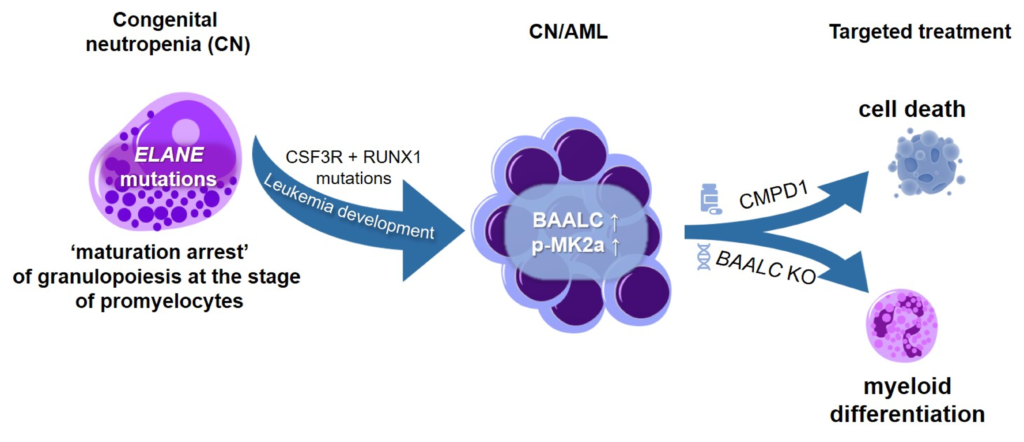
CN is a pre-leukemia syndrome with a cumulative incidence of myelodysplastic syndrome (MDS) or acute myeloid leukemia (AML) of more than 20% after 20 years. The progression of CN to MSD and/or AML is a multistep process. HSPCs harboring inherited mutations acquire early driver mutations during neutropenic phase (e.g., CSF3R mutations), followed by additional mutations before onset of full-blown MDS/AML.
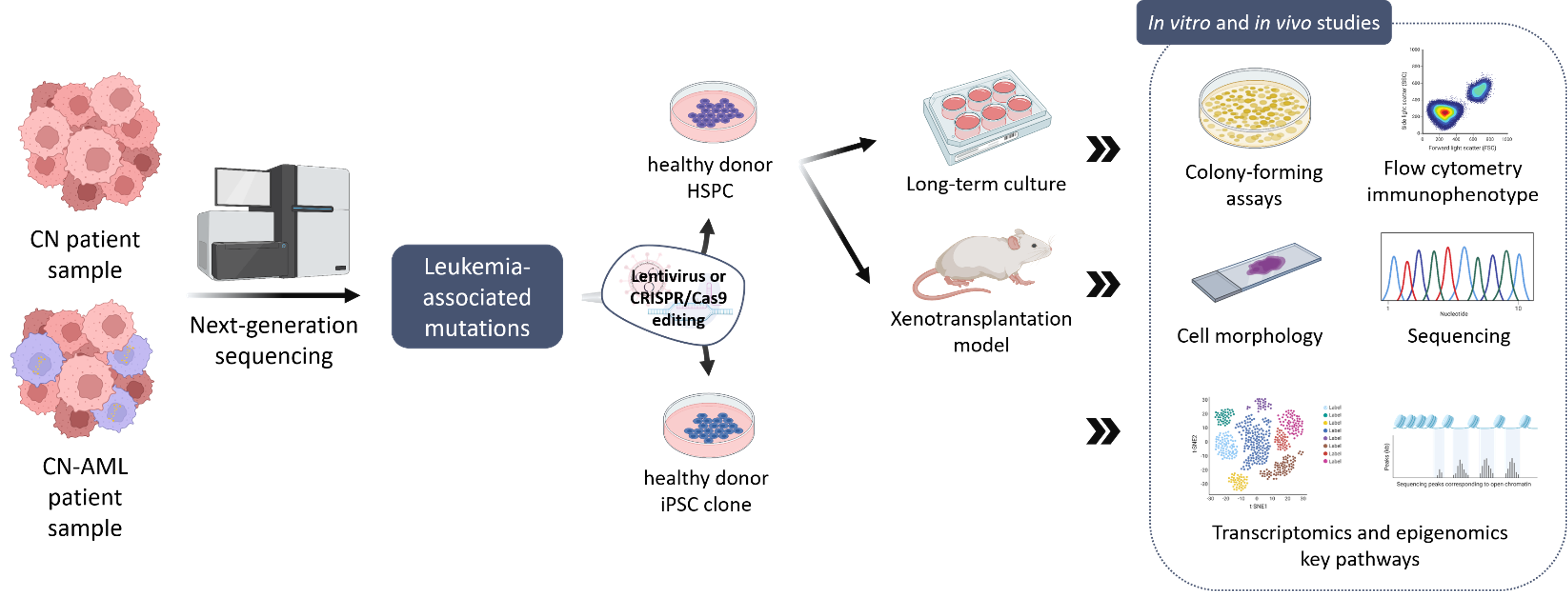
In order to improve early diagnosis and treatment of CN patients that may develop leukemia, our group aims to identify additional gene mutations in leukemia-associated genes in CN-MDS/AML patients using a NGS-based sequencing panel of leukemia-associated genes and comparing CN/AML patient samples to CN samples in order to identify additional candidate mutations. To further characterize these leukemia-associated mutations in CN-MDS/AML patients, we use different experimental models to assess the leukemogenic potential of new mutations in malignant transformation and uncover intracellular signaling pathways involved in disease development.
iPSC disease modeling of stepwise leukemogenesis in severe congenital neutropenia
Our group established a disease model of stepwise leukemogenesis in CN/AML by CRISPR-Cas9 gene editing of CN/AML patient-derived iPSCs and subsequent hematopoietic differentiation. This model we also used for AML drug discovery. We identified BAALC upregulation and the following phosphorylation of MK2a as a key leukemogenic event. BAALC deletion or treatment with CMPD1, a selective inhibitor of MK2a phosphorylation, blocked proliferation and induced differentiation of primary CN/AML blasts and CN/AML iPSC-derived hematopoietic stem and progenitor cells (HSPCs) without affecting healthy donor or CN iPSC-derived HSPCs. Our disease model is suitable for future investigation of stepwise leukemogenesis and AML drug screening. This study suggests that targeting BAALC and/or MK2a phosphorylation may prevent leukemogenic transformation or eliminate AML blasts in CN/AML and RUNX1-mutant BAALChigh de novo AML (Dannenmann B, Klimiankou M et al. Cell Stem Cell. 2021).
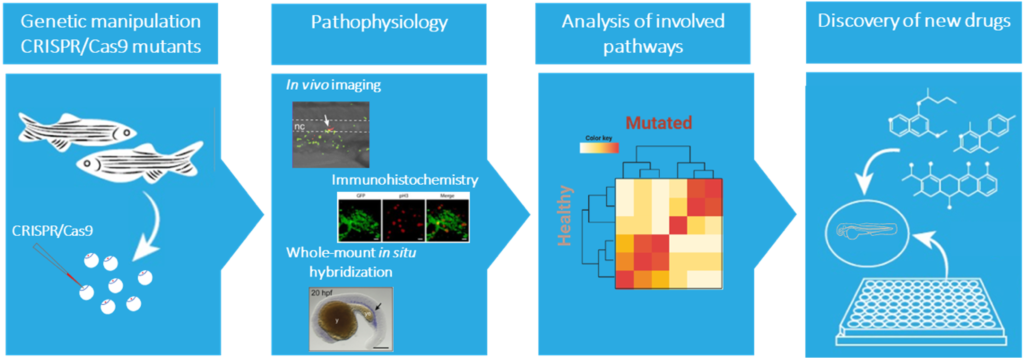
Zebrafish models
The information acquired from model organisms is essential for understanding the development of a disease. The zebrafish (Danio rerio) is an attractive complementary model system. The molecular and cellular mechanism underlying hematopoiesis are largely conserved between zebrafish and humans, making the zebrafish a very suitable vertebrate model to study normal and malignant blood development. Also, the optical transparency and ex vivo development of the embryos and larvae allows monitoring throughout embryogenesis, providing unique accessibility to embryonic lethal mutations. Additionally, the availability of fluorescent transgenic reporter lines allows an easy observation of different cell behavior and analysis of niche interactions.
Our current knowledge on genes associated with severe congenital neutropenia is predominantly derived from clinical observations and in vitro studies. In our group we use zebrafish as a model to fill the gap and understand the molecular mechanisms underlying congenital neutropenia and leukemia development. In addition, the zebrafish will serve as an in vivo platform to identify new avenues for developing tailored therapeutic strategies for patients for various types of blood malignancies, including specific subtypes of ALL, AML, MDS, and MM as well as inherited pre-leukemic bone marrow failure syndromes.